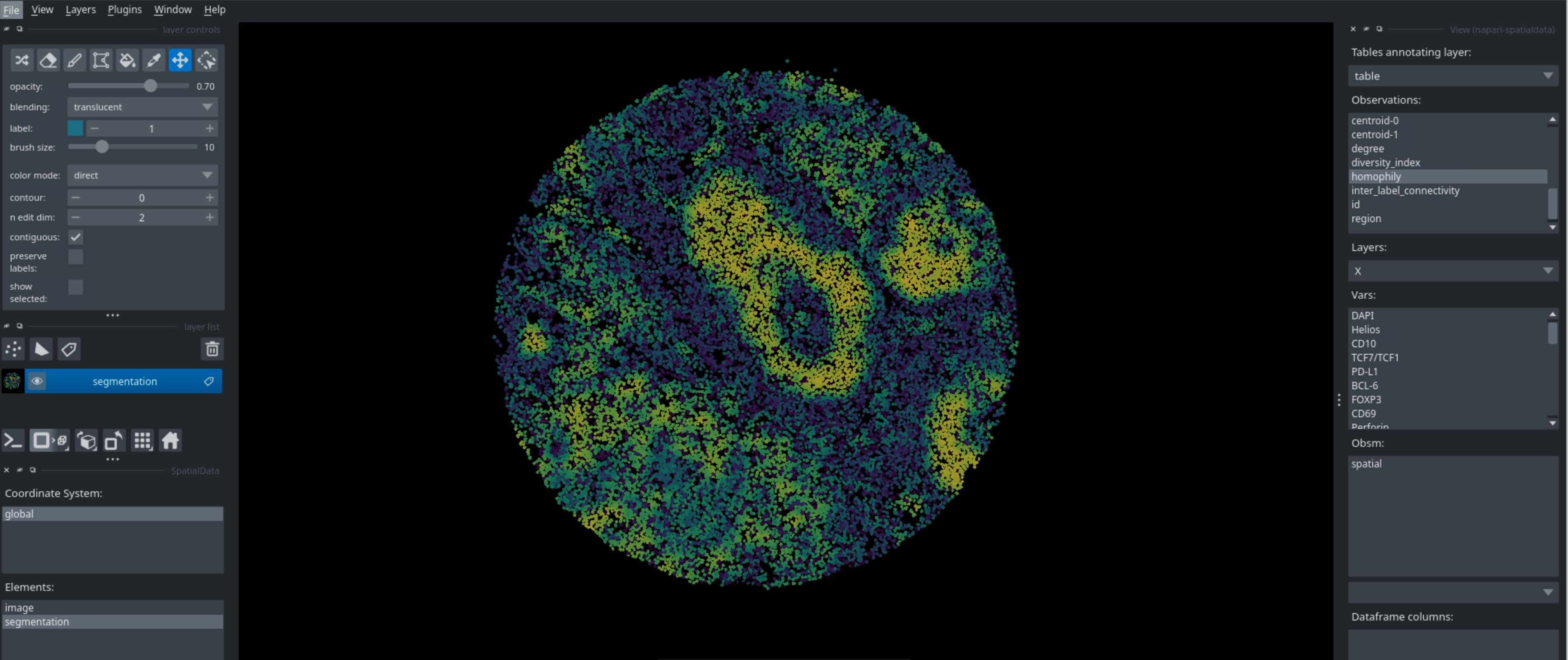
Interactivity with napari#
Spatialproteomics has a variety of functions to ensure interoperability with existing tools. For example, we can export the spatialproteomics object into spatialdata, and the use napari for interactive visualizations.
If you want to follow along with this tutorial, you can download the data here.
[1]:
import spatialproteomics as sp
import xarray as xr
from napari_spatialdata import Interactive
import numpy as np
/home/meyerben/meyerben/.conda/envs/tmp_env_3/lib/python3.10/site-packages/dask/dataframe/__init__.py:31: FutureWarning: The legacy Dask DataFrame implementation is deprecated and will be removed in a future version. Set the configuration option `dataframe.query-planning` to `True` or None to enable the new Dask Dataframe implementation and silence this warning.
warnings.warn(
No OpenGL_accelerate module loaded: No module named 'OpenGL_accelerate'
[3]:
# loading in a data set and performing some formatting for convenience
ds = xr.open_zarr("../../data/LN_11_1.zarr")
[4]:
# turning the spatialproteomics object into a spatialdata object
sd_obj = ds.tl.convert_to_spatialdata()
sd_obj
INFO Transposing `data` of type: <class 'dask.array.core.Array'> to ('c', 'y', 'x').
INFO Transposing `data` of type: <class 'dask.array.core.Array'> to ('y', 'x').
[4]:
SpatialData object
├── Images
│ └── 'image': DataArray[cyx] (56, 3000, 3000)
├── Labels
│ └── 'segmentation': DataArray[yx] (3000, 3000)
└── Tables
└── 'table': AnnData (16871, 56)
with coordinate systems:
▸ 'global', with elements:
image (Images), segmentation (Labels)
[ ]:
interactive = Interactive(sd_obj)
interactive.run()
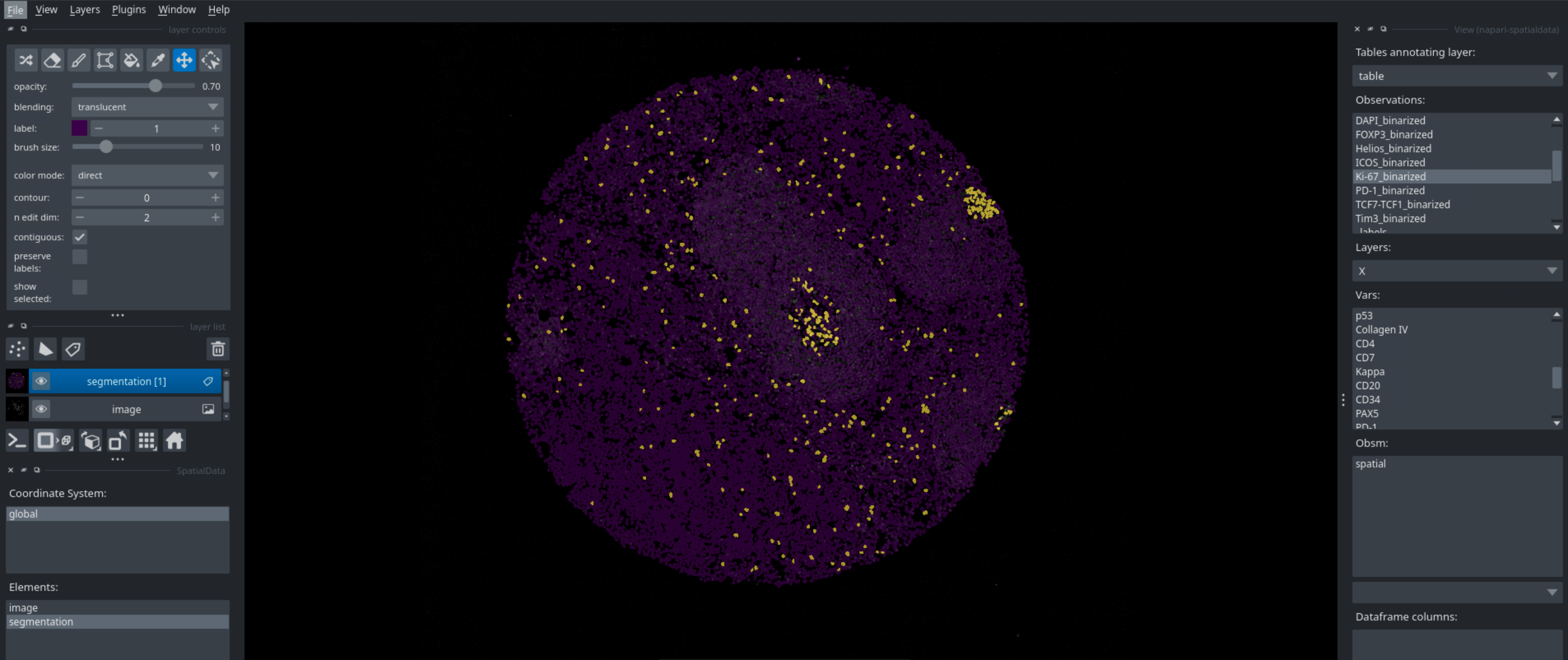
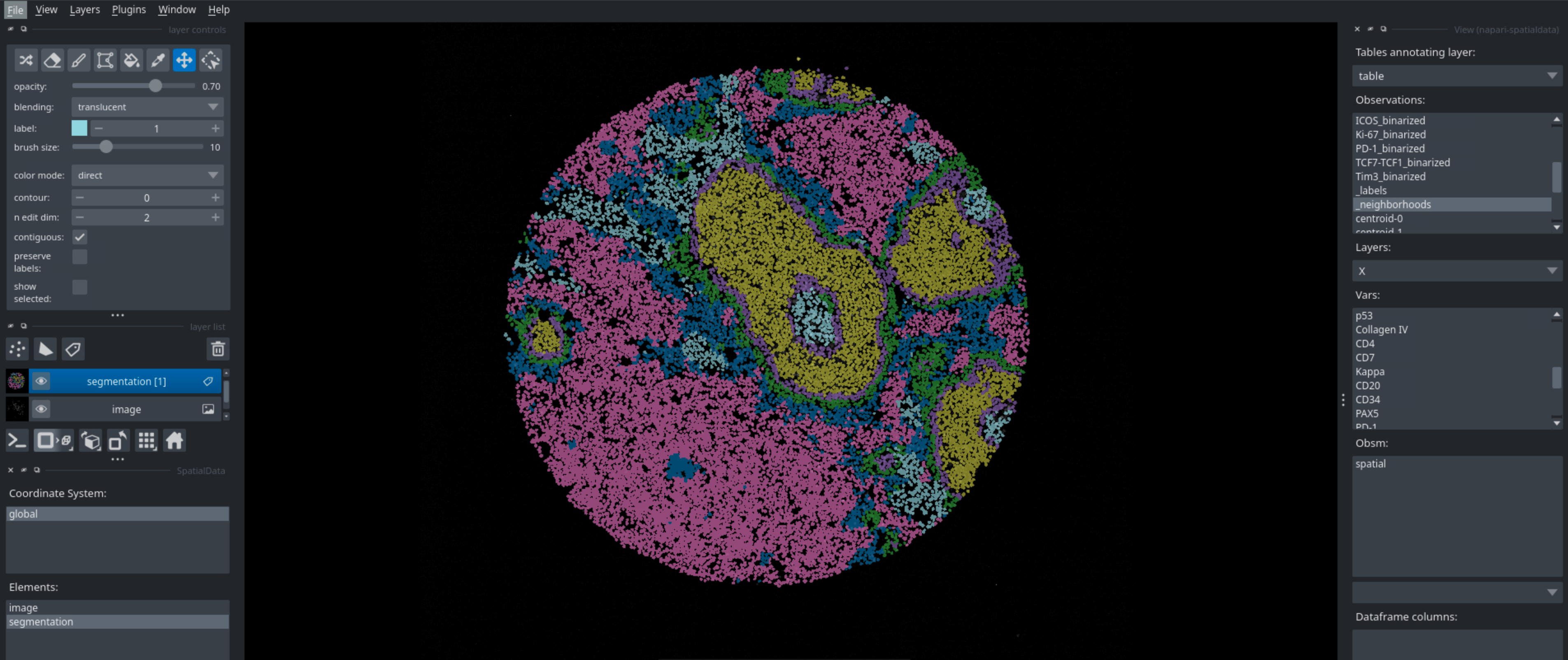
Running this code in a python script will open napari, which you can then use to interactively look at your data. You can look at different markers, visualize cell types, neighborhoods, and many more. Below are some examples of what you can visualize with napari.
Markers#
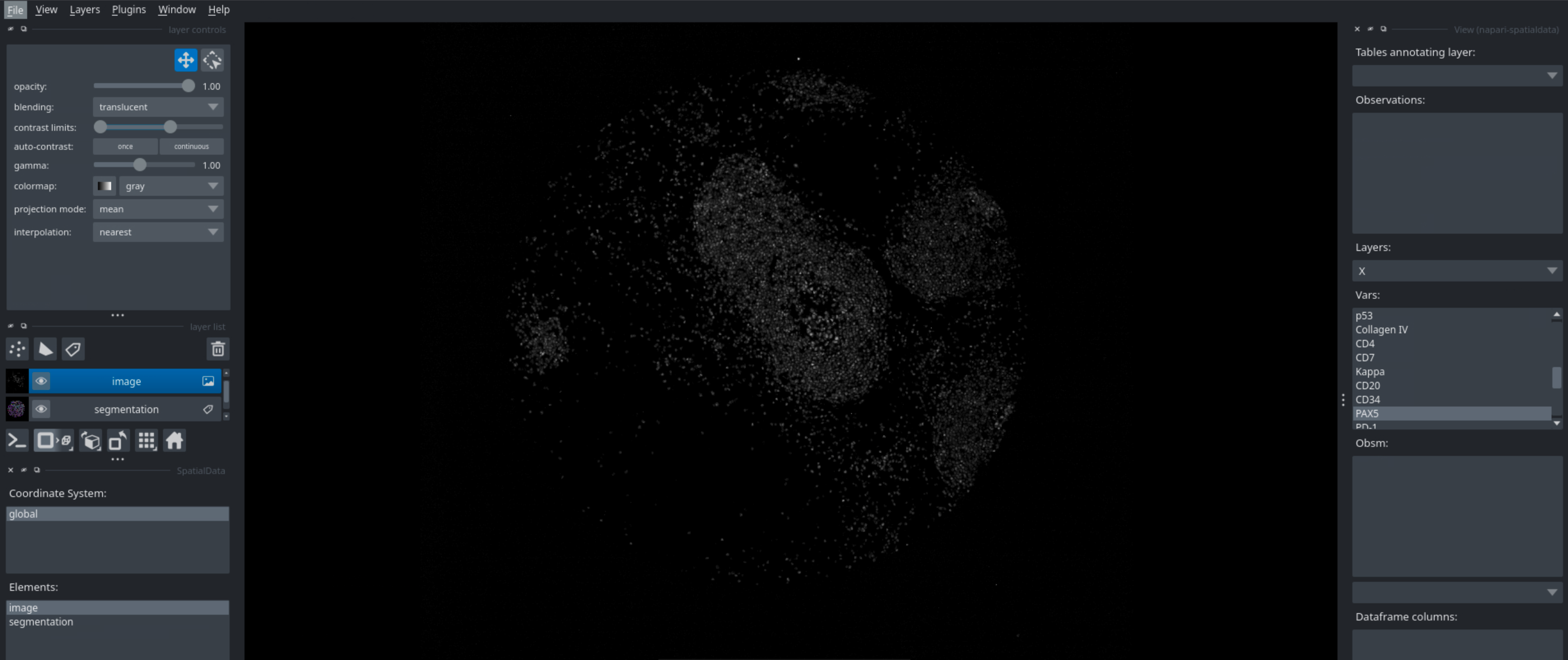
Segmentation#
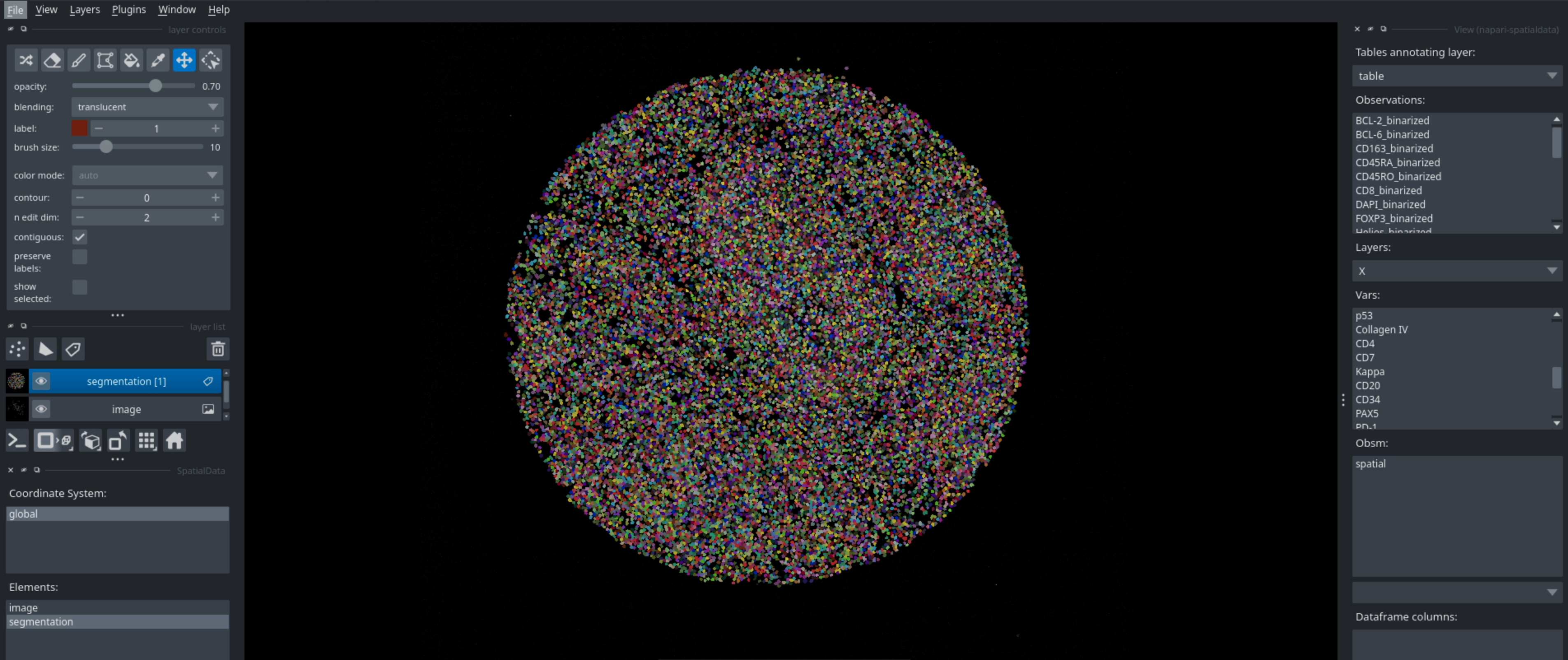
Cell Type Labels#
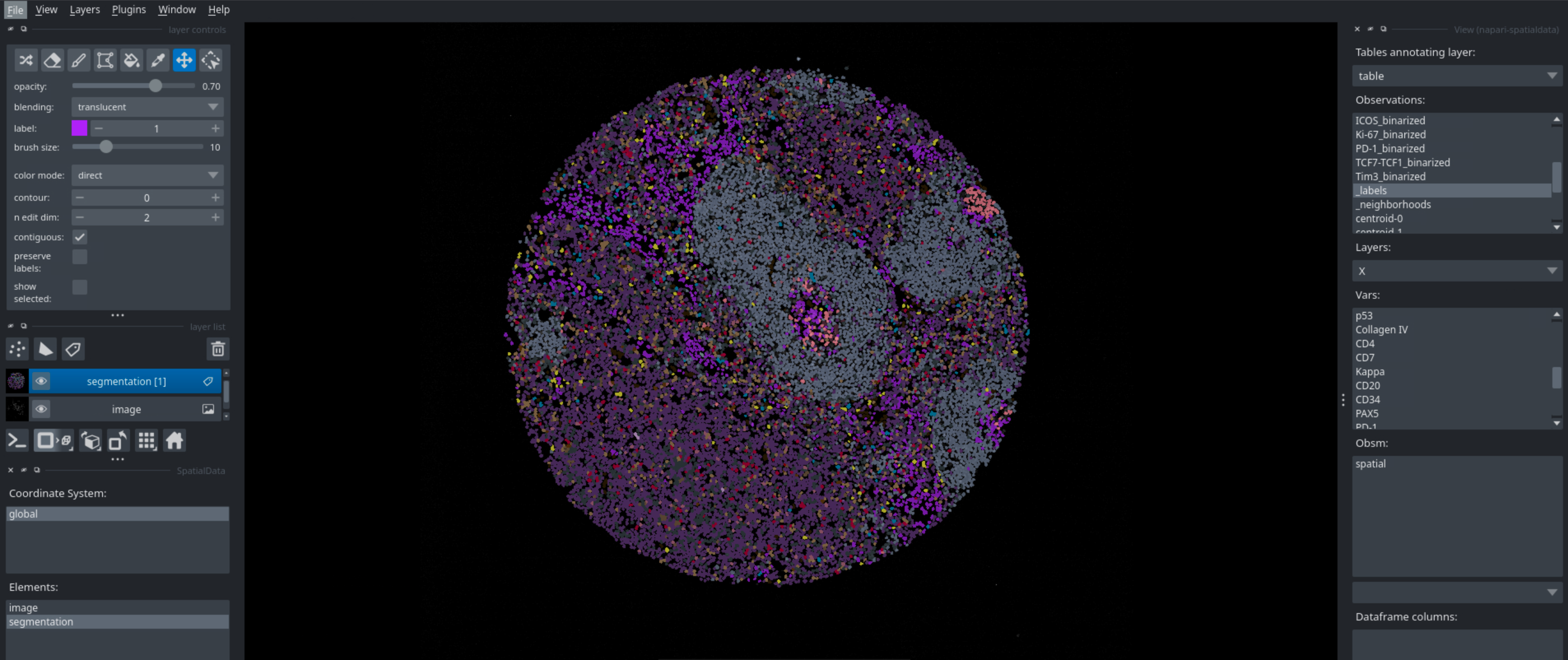
Binarization#
Neighborhoods#
Features#